Part 2: Patent Busting A Literature-based Approach
July 28th, 2025 By John Widen
Part 1 of this patent busting blog series discusses how to search for
patent applications and literature to find relevant chemical matter for your protein target of interest. Finding relevant chemical matter
can be challenging in more ways than one. Sometimes, there aren’t a lot of great starting points for a target. So, you have to think about
whether that chemical matter is good enough to start a program or if some sort of screen is beneficial. On the other hand, for really popular
targets like KRAS it can be intimidating because there are SO MANY patent applications that are all related to each other,
covering each other’s gaps in IP. It can also be tough to determine if the chemical matter actually directly binds to your target of
interest or if it interacts with a different protein that modulates it. To find this out typically requires quite a bit of investigatory work.
In this article, I’m going to discuss the different sections of a patent application and some aspects of BUSTING!
I will use a relatively new patent application (WO2024089155) from Boehringer Ingelheim (BI) that stakes out claims on
STING (Stimulator of Interferon Genes) antagonists. I’m using this application because it currently stands on its own in
terms of the first to publish this chemical matter as a STING antagonist. There are many other applications that involve
STING agonists because this mechanism is being investigated to inhibit cancer growth and progression. BI has a STING
agonist program listed on their website but not antagonist. If I were a betting person, I would guess that the group working on
small molecule STING agonists stumbled upon antagonists and decided to start a program focused on that because there are plenty
of indications where this could be useful.
I’m not going to go deep into the history and biology of STING in this article. I am using this application
as an example of what to look for when patent BUSTING. I am going to discuss the different sections of a patent application
and the information that can be utilized to evaluate the target of interest and the chemical matter.
One more note on downloading PDFs for patent applications. Ctrl+F search does not work on standard patent application
PDFs because they are scanned or encoded. SciFinder and others offer a “PDF+” for download, which you should choose that option when available.
Those PDFs have searchable text. Otherwise, you have to use other tools that use optical character recognition (OCR) if you want to search
for words and phrases.
That’s enough lollygagging. Let’s start to go through WO2024089155 from BI and see how we can BUST this patent.
There are several sections of a patent application that are important when evaluating chemical matter and the target of interest.
I'll go through and discuss some aspects of each of them below.
Abstract and introduction:
You can usually find the Markush structure here. If not, I would scroll down to the claims to make sure the patent
application is not a methods or use case as discussed in part one. No Markush structure anywhere in the patent means it’s
either a selection case or doesn’t contain any novel chemical matter. These sections also include a background on the target
of interest and related disease indications. The indications are usually very general and all encompassing but can still be
useful to look at it if you're pitching a new target to enter into your company portfolio. If anything, it provides some
keywords to search when thinking about demonstrating on target modulation and efficacy in pre-clinical models, which all
ties back to the disease indication(s) of interest.
A screen shot of the abstract is below as an example.
Prior Art:
This is usually listed right after the introduction and also in a search report all
the way at the end of the document. I like looking at applications from the World Intellectual Property Organization
(WIPO) so it is called the “International Search Report” for WO applications. These two sections will point to any
relevant patent applications and publications, which include patents that might be in the same chemical matter class
or any published structures. It can also tell you if a patent application’s claims likely infringes on other patent
application claims. It is always good to look through these as you dig deeper.
Claims:
This is right after the background and sometimes the definitions
(I’ll get to the definitions next). This is where the patent BUSTING happens.
The goal of the claims is to destroy the novelty of the chemical matter presented in the
application. In other words, claims are framed by the example molecules that are presented
in the patent application and want to cover as much chemical space as possible without infringing
on any other prior art (another legal definition). Prior art is the lingo for other claims and chemical
structures that have been published in any other patent application within twenty years. There are some
nuances, but I believe this to be true most of the time. So, any patent application will try to expand
the claims (i.e. chemical matter covered) as much as possible surrounding the examples presented.
The idea is that even though the examples don’t include every possible iteration of molecules that
could be active the claims cover obvious changes that would still likely have activity
against the target of interest.
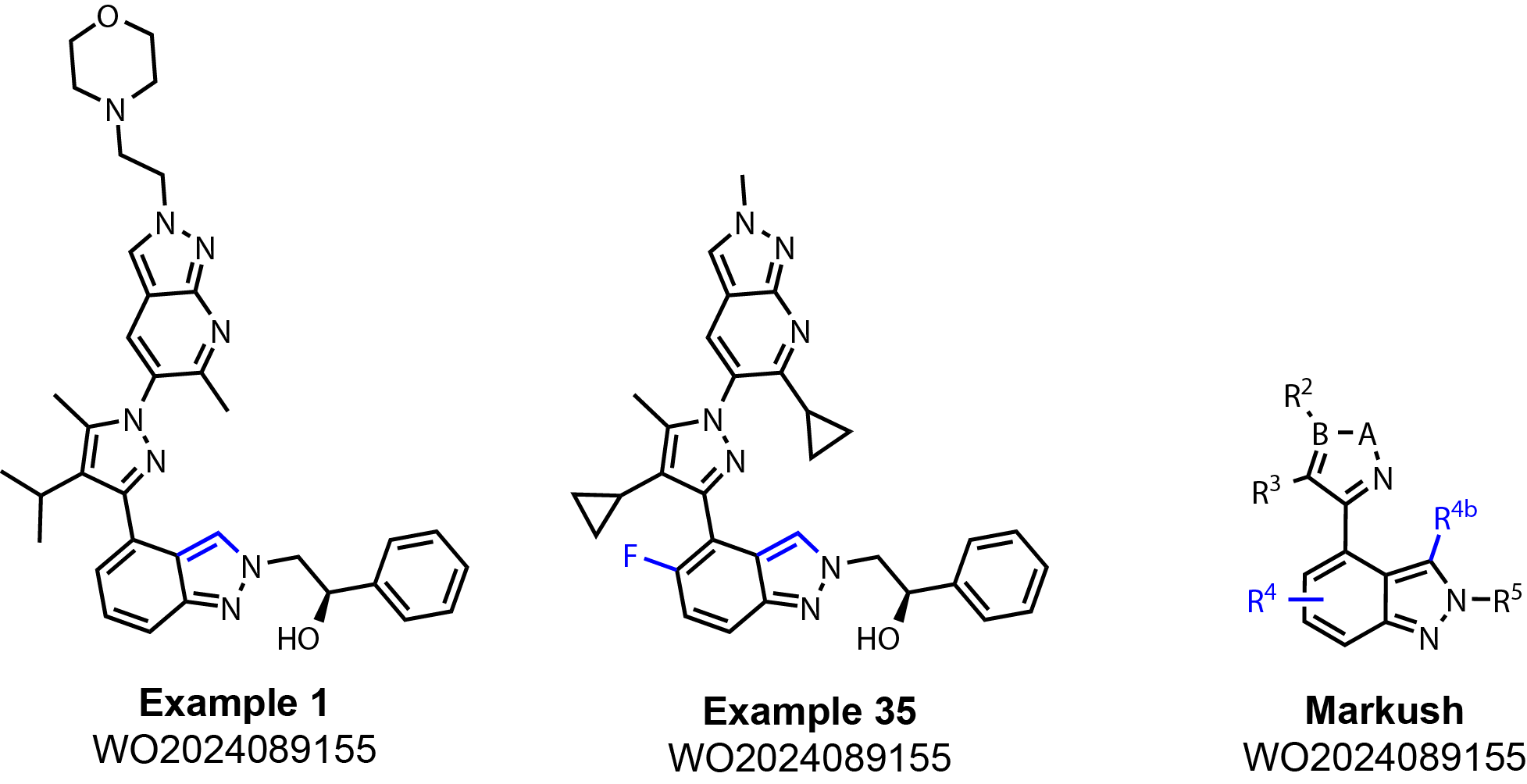
This is where an example comes in handy. Below are structures of
Example 1 & 35 in WO2024089155 on page 38 & 50 respectively next to the Markush
structure (Fig. 2). (Side note: Always take note of the pages
where different sections start. It’ll save you time when you inevitably have to scroll back and forth.
Additionally, having at least two of the same PDF or making a split view of the document also helps.
I’ll get into annotating patents later.) The first set of claims start on page 3.
Let’s focus on R4 and R4b of the Markush structure.
The claims define R4 and R4b as follows:
- R4 is H-, F-, or HO-;
- R4b is H-, F-, Cl-, Br-, NC-, or HO-
Example 35 is the ONLY example out of 113 that contain a
fluorine on the bottom indazole (highlighted in blue). BUT, the
claims based on the Markush structure cover fluorine AND hydroxyl (OH-)
substitution at any position on the indazole. They only made ONE compound
and then extended to now many different possible combinations in the claims!
Right away this is a little weird because why didn’t they claim other
halogens such as chlorine, bromine, and iodine? Additionally, why did they not claim other
substituents about this ring?
There could be several answers
including that they are challenging to synthesize, chemical space is covered in another patent application making it
prior art, or those compounds are inactive (or very likely to be inactive). Either
way it’s the first identification of a GAP in the claims! Write that one down.
Maybe they just decided to reduce the claims here. There’s only one way to find
out if these substitions are tolerated and that is to make derivatives that exploits this gap. The question is
whether these types of substitutions provide an advantage to the properties of
the molecule. It is the first point of differentiation that could be exploited.
Another example is in R4b. This claim does include common halogens in
addition to hydroxyl and a nitrile (CN-). BUT, if you look through all 133 examples in the
patent application there is NOT A SINGLE example with a substitution other than hydrogen
(highlighted in blue). Not a single example! Yet these claims are covering that chemical space.
Another odd choice. Now this claim may get limited by the patent office after review and conversion
to a patent, but there is no way of knowing that right now and would take multiple years to determine.
So, for now the assumption has to be that the claim will stick. It would be a significant risk to try
and exploit that pseudo-GAP. It’s not that juicy anyways so we will let it go.
Those are two examples of how claims can cover additional chemical space beyond the
examples shown in the patent. There are plenty of other things to look at in the claims to identify GAPS.
But I will stop there for now and comeback to these claims in another post. I’ll go into finer
details on evaluating these claims and looking for omissions with more examples. Moving on to the next section!
Definitions:
This section comes either before or after the claims. In the case of WO2024089155 the
definitions section is after the claims and examples (tricky!). Definitions are exactly as
the name suggests. They define chemical terms inside the claims. Some patent applications
covering novel chemical matter don’t have definitions and are incorporated in the claims,
but I would say most have a separate section. It might seem arbitrary to define chemistry
terms but it really isn’t. Again, I’ll use a couple of examples to explain this point and how
looking through the definitions when patent BUSTING is very important.
Usually, the most important definitions are for terms such as alkyl, alkylidene,
carbocyclyl, cycloalkyl, heterocyclyl, aryl, and heteroaryl. There are specific things
l like to look for inside the definitions that could provide ways to BUST.
For instance, looking through the definition of carbocyclyl (a.k.a. cycloalkyl)
the definition includes bridged systems, spiro systems, and unsaturations. This also applies to other
definition terms mentioned above. If you find these missing from the definitions, you might have
found a way to BUST this patent. It is an easy place to check for GAPS in the claims.
Aryl definitions can also leave out specific terms such as multi-ring systems that include
five-membered rings. Alkyl and alkylidene definitions can leave out substitutions of halogens
or other functional groups. In the case of this patent application, these definitions are included
and overall don’t leave any obvious omissions upon first inspection. So, good for them.
BUT WAIT! One glaring omission from the claims or definitions in WO2024089155 is ISOTOPES.
That is a pretty big mistake unless it is covered elsewhere in a future application or
previous application. Medicinal chemists incorporate deuterium isotopes into molecules
to potentially reduce metabolism. It is also a strategy to get around IP. Read more HERE.
However, this may not be as much of an issue anymore because of a
recent court ruling.
I’ll try to keep this short and say
several companies have
taken advantage of GAPS in
claims where isotopes are not mentioned.
Teva gained FDA approval for the first drug containing
deuterium in 2017 in which they took an already approved drug and incorporated deuterium atoms.
Companies have been founded on the premise of looking through patent
literature and identifying compounds where deuterium incorporation can improve metabolism and lead to better drugs.
You can read more by following the links in this paragraph.
Most applications now cover different isotopes with a simple phrase in the definition
or claims mentioning that any molecule or claim shown can also include any and all isotopes of an atom.
Many applications actually go out of their way to exemplify incorporating deuterium into their molecules.
You never know if deuterium will save a molecule from getting demolished by CYP enzymes. I’ve certainly
observed fairly drastic improvements in liver microsome clearance values between matched pairs, but far more
often deuterium doesn’t work that well. Either way, if you are writing a patent it is a very easy thing to
incorporate to cover your bases, but now this method of patent busting might be considered obvious.
Either way, it is something to look out for.
Examples:
There is usually a table of the example molecules for which the claims are based on (a.k.a. picture claims).
I’ll mention that there are annotation services that will manually convert these images into structural data and
associate the examples with the biological activity in the patent application. It is typically delivered as an
SDF file that can be opened with medicinal chemistry software such as Vortex (by Dotmatics), Data Warrior (free),
and others. These annotations are typically done for a few dollars a compound. It can be expensive and can take a
while but it makes things SO MUCH EASIER. If you have the resources and it is a relevant application I would
suggest doing that. Otherwise, this has to be done manually in some way where you can identify the molecules
that are most potent that you might want to synthesize for initial evaluation, setting up assays, etc.
The exemplified molecules are typically what was synthesized and tested.
But alas, companies also put in prophetic molecules. These are picture claims of
molecules but as the name suggests were not synthesized; ONLY THOUGHT OF. I personally think
this is an annoying trick that shouldn’t be allowed. But it happens and it is another method on top of the claims to exclude others
from making specific compounds that may have activity against the target of interest and good properties.
I think the only risk of including prophetic examples in a patent is that if one of them becomes the FDA
approved drug than the time for having IP protection during commercialization will be shorter. The company
has the 20 year timeline started before even making and testing the molecule, which takes quite a bit of time.
It eats into your window of opportunity for exclusivity in the market to make as much money as possible.
This is a relatively low risk so is quite common.
Synthetic Methods and Preparation:
This is the section where the invention is described (i.e. the syntheses).
The author of the patent application has to provide enough detail that someone ‘skilled in the art’
could synthesize the molecules. It usually provides general schemes used to synthesize the molecules
and specific synthetic methods for the example compounds.
There is also characterization data for each molecule to demonstrate that the actual compound was
synthesized and tested. The minimum requirement is mass spectrometry data but many applications include H1 NMR data.
Biological Assay Data:
This section (page 222) is usually at the end and contains information about the assays
used to characterize the compounds and tables that include the assay data demonstrating activity.
It is pretty rare for authors to include actual numbers in the tables. Instead, the activity will
be put into potency buckets where letters (A-D) or a series of plus signs (e.g. ++++ to +) represent the potency of compounds.
This usually destroys any chance of someone BUSTING the patent to pick out the best molecule or
derive meaningful SAR. The potency buckets are usually sufficiently large to eliminate the ability to
determine if certain substituents lead to significant gains in potency.
The assay information is useful when determining the relevance of the patent upfront.
It’s best when an assay is presented that demonstrates direct interaction between the molecules
and the protein of interest. This is typically in the form of a biochemical assay. Otherwise, if
it is a phenotypic assay and there isn’t precedent for the molecules directly interacting with the
protein of interest, then you should be weary of the chemical matter and its relevance to the program.
Fortunately, WO2024089155 reports data for an HTRF binding assay and a whole blood assay.
It also reports actual numbers for both assays! That’s pretty awesome when that happens because
it makes it much easier to determine SAR and decide which molecules to make for initial evaluation.
When getting a project started, the description of the assays are useful as well. Patents are not peer
reviewed but again it has to provide detail for those skilled in the art to evaluate the compounds.
This is usually a good place to look when thinking about primary assays for evaluating novel chemical
matter and what will be used to drive SAR.
I think we covered the main sections of a small molecule-based patent application! PHEW!
This blog post is already way too long. I will stop there and pick it back up in what would be
PART 3 of this blog series. Next time I will go through the claims in more detail and identify the
GAPS! Stay tuned and thanks for stopping by.
The site does not have a comments section yet! Hopefully, very soon! Until then please drop me a line at jwiden@chemjam.com.
If you provide comments on my articles I reserve the right to post them on this website as additional commentary. My goal is to have an open discussion!