Orthosteric versus allosteric SARM1 inhibitors
August 4th, 2025 By John Widen
Nura Bio is a biotech company in South San Francisco focused on developing ‘neuroprotective medicines.’
In 2020, Nura Bio announced a Series A raise of $73 million that apparently did not close until 2024 when they
announced raising an additional $68 million over that four-year time period. Nura Bio has two publicly known
small molecule programs based on patent application filings and publications: SARM1 (Sterile alpha and
Toll/interleukin-1 receptor motif-containing 1) and TREM2 (Triggering Receptor Expressed on Myeloid Cells 2).
I’m going to focus on SARM1 as a drug target for neurodegenerative diseases in this article. However,
I will mention briefly that TREM2 as a drug target for Alzheimer’s disease is certainly being questioned
after a Phase 2 clinical failure of a monoclonal TREM2 antibody AL002 put forth by Alector in partnership
with Abbvie. Among other clinical failures for TREM2-targeting antibodies. That is for a different discussion. So, on to SARM1!
There are four patent application filings by Nuro Bio
(WO2022031736, WO2022060812, WO2023009663, and WO2024026368) covering chemical matter of orthosteric
inhibitors of SARM1, a nicotinamide adenine dinucleotide (NAD+) hydrolase. SARM1 is abundantly
expressed in neurons and is a drug target because of its role in axonal degeneration observed in multiple
sclerosis, amyotrophic lateral sclerosis (ALS), chemotherapy-induced peripheral neuropathy (CIPN),
diabetic neuropathy, and other neurodegenerative diseases. During neuronal damage SARM1 is activated
by an exchange of NAD+ by nicotinamide mononucleotide (NMN) in a SARM1 allosteric binding site.
Activation of SARM1 NADase activity triggers an energy crisis and propagates downstream signals leading to
neuron degeneration. Therefore, small molecule inhibition of SARM1 is desirable to prevent neuron degeneration.
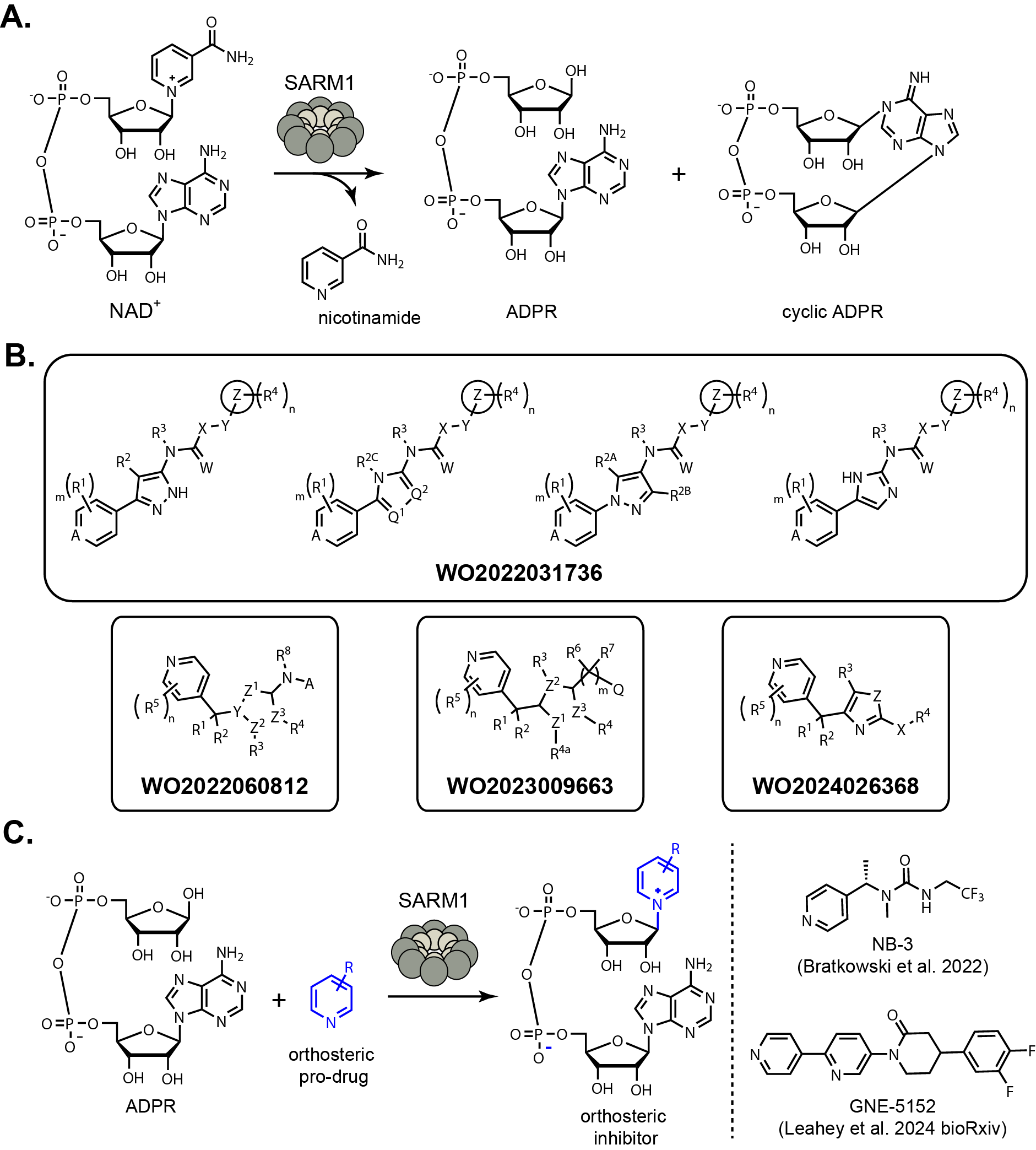
There have been two main approaches to inhibiting SARM1. The first approach is using orthosteric
(active site) inhibitors that act as a substrate for SARM1. The natural SARM1 catalytic activity cleaves
NAD+ into nicotinamide, adenosine diphosphate ribose (ADPR), and cyclic ADPR (Fig. 1A). The Markush
structure from each patent application covering orthosteric inhibitors of SARM1 from Nura Bio are
below (Fig. 1B). The pyridyl nitrogen is required for activity of these inhibitors
because it reacts with ADPR (replacing nicotinamide) forming an adduct that is the active site inhibitor (Fig. 1C).
Orthosteric inhibitors such as NB-3 and GNE-5152 are actually prodrugs because they undergo a reaction to produce the active molecule
that inhibits SARM1. Pretty interesting!
Nura Bio published their work developing orthosteric inhibitors of SARM1 demonstrating this
mechanism in in 2022.
Their orthosteric SARM1 clinical candidate NB-4746 completed a
phase 1 safety trial in Australia (registration number:
ACTRN12623000476639).
The issue with these inhibitors is that they cause paradoxical activation of SARM1
NADase activity at sub-inhibitory concentrations exacerbating neuronal degeneration.
Several publications from F. Hoffmann-La Roche
(Mani et al. 2025) and Genentech
(owned by Roche, Leahey et al. 2024)
demonstrate this paradoxical activation of SARM1 in several different cell-based experiments and in vivo
models of neuron degeneration. I am familiar with one other industry group that also discovered this issue with orthosteric SARM1 prodrugs
but did not publish their findings. So, that is three independent research groups that have
identified this issue with orthosteric SARM1 prodrugs that could have major negative clinical impacts.
Genentech and Roche demonstrate that completely covering the target (>EC95, >30 mg/kg in mice)
results in reduction in nFL (neurofilament light chain), which is a biomarker for neuron degeneration.
However, at lower doses the nFL increases significantly compared to DMSO control.
These lower doses also caused acute adverse events including death. Genentech also saw adverse events
at the highest dose tested of 100 mg/kg for an orthosteric SARM1 inhibitor. These publications
demonstrated that this exacerbation was SARM1 dependent and only happened in conjunction with neuronal injury.
This sub-inhibitory activation of SARM1 is a general phenomenon for orthosteric inhibitors as many structurally distinct
molecules were evaluated. Additionally, there is a ceiling effect in cell-based
and biochemical assays commonly used to drive SAR. This means that high concentrations of NMN (200 µM)
that are used for these assays causes complete activation of SARM1. So, an orthosteric inhibitor that
causes paradoxical additional activation would be missed because there is no more SARM1 to be activated.
For this reason, one can imagine how this effect might be missed.
SARM1 is an octameric protein with eight allosteric binding sites for NAD+/NMN that
undergoes a large conformational shift upon exchange of NAD+ for NMN. Activation of SARM1
forms six active sites that are capable of NADase activity. Although the authors did not fully elucidate
the mechanism of paradoxical activation, they spoke to several possibilities including sub-inhibitory
concentrations of inhibitor causing increased activity at other sites and forming oligomers that
activate inactive SARM1 complexes. A
separate publication
from Genentech goes into additional detail in support of the oligomerization mechanism.
Nura Bio has a phase 1b clinical trial registered in Australia (RN: ACTRN12624001072505p)
to follow up on the phase 1 trial for NB-4746 for what appears to be a potential CYP induction issue.
This would suggest they are marching on with their orthosteric SARM1 inhibitor.
These findings by Roche and Genentech would suggest a very significant risk for treating
patients with an orthosteric SARM1 prodrug including exacerbating neuronal degeneration instead
of slowing it down. Overcoming this risk by dosing to consistently cover >EC95 in
humans seems like a risky endeavor and would very likely lead to causing more harm to patients
than good. I think in light of this new information any clinical trials surrounding this mechanism
should probably be stopped or at the very least adjusted to monitor patients closely.
A second approach that has been taken to inhibit SARM1 involves targeting an allosteric site.
Indeed, there are patent applications from several companies including Nura Bio that focus on
allosteric inhibition of SARM1 that avoids paradoxical activation. Nura Bio published a patent
application (WO2025090514) in May. They must have gotten some sort of hint that orthosteric
inhibition of SARM1 carried a risk of causing increased neuronal damage. Other companies that
have patent applications covering allosteric inhibitors of SARM1 include
Tenvie Therapeutics
(a spin out from Denali Therapeutics) and Sironax.
I’m sure Genentech is developing SARM1 allosteric inhibitors but have yet to have any published patent applications.
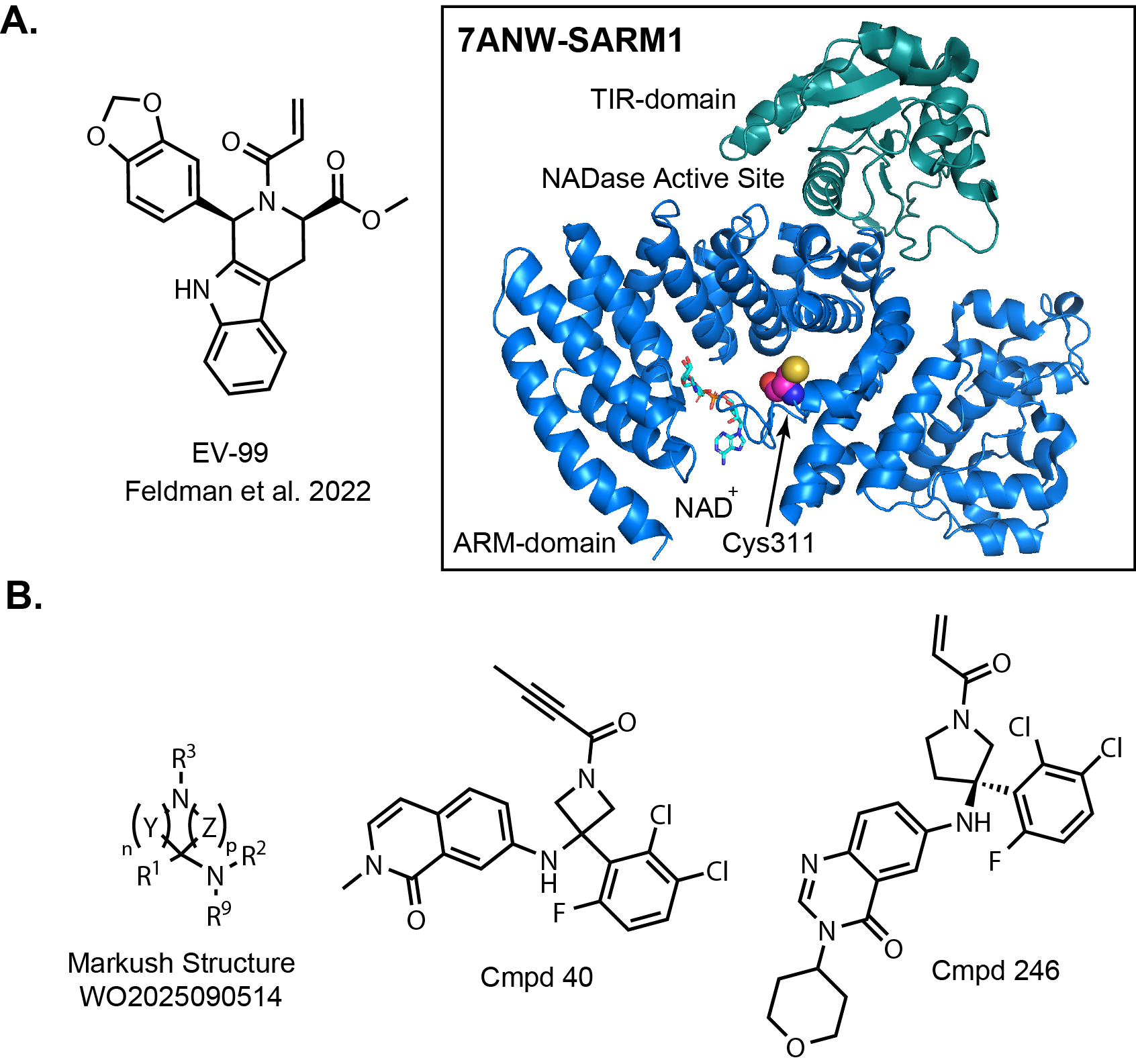
I was surprised to see that Nura Bio’s patent application focused on covalent inhibitors!
If anyone knows me, they’ll know that I like covalent inhibitors. The molecules are likely based
on a publication by a group at Scripps that includes Stu Schreiber and Ben Cravatt.
They identified an acrylamide covalent inhibitor of SARM1 while screening a complimentary
stereochemical covalent library using LC-MS/MS chemoproteomics. The idea of these libraries
is that each molecule is a single stereoisomer with the opposite stereoisomer(s) also present
in the library. Each molecule has both sets of enantiomers or in the case of diastereomers two
to four sets of molecules depending on symmetry. In this way, identifying single isomers with
activity versus their counterparts that lack activity identifies molecules that are engaging binding
sites in a specific manner. It’s a nice control to give confidence that the interaction between the
ligand and protein are specific and therefore can be optimized.
The identified active isomer EV-99 was demonstrated to engage C311, which is located in the
ARM domain of SARM1 (Fig. 2A). This residue is on a flexible loop located adjacent to the
allosteric NAD+/NMN-binding site. Nura Bio has different but related molecules in their
patent application. Their Markush structure leaves a lot to the imagination but there are two examples
that are in the highest potency bucket to provide a better idea of their chemical matter in Fig. 2B below.
R3 is the covalent warhead off of the nitrogen of a central saturated ring. Pyrrolidine is the
most prevalent structure represented in the examples. My guess is that they homed in on that scaffold.
Otherwise, kudos to the team for providing a meaningless, single time point assay with large potency
bucket ranges to demonstrate activity against SARM1.
One of my biggest pet peeves is characterization of covalent inhibitors using a
single time point assay. Covalent inhibitors are by nature time-dependent and thus
characterizing their reactivity by determining kinetic parameters such as Kinact,
Ki, or Kinact/Ki when appropriate is necessary to understand
potency and reactivity. A patent application
would not provide that information because the only goal is to demonstrate activity. Giving any other information away
like the kinetic parameters of each molecule would not be beneficial to them.
It'll be interesting to see in the future if anyone else is in the covalent chemical
matter space for SARM1 inhibitors to compete with Nura Bio or if they are going it alone.
I’m sure there are still quite a few medicinal chemists that cringe when they hear the mention of a covalent molecule.
The old ways of thinking, which lack real world evidence,
are that covalent molecules have increased toxicity risks. I would encourage everyone that
thinks that to read this review
and this review and
this review and this
review.
I’ll stop there and thanks for reading.
The site does not have a comments section yet! Hopefully, very soon! Until then please drop me a line at jwiden@chemjam.com.
If you provide comments on my articles I reserve the right to post them on this website as additional commentary. My goal is to have an open discussion!